Ambiscript is a functional DNA notation that supports the manipulation and analysis of genetic data
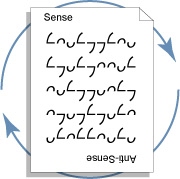
Ambiscript uses character symmetries to facilitate the complementation of deoxyribonucleic acids (DNA) and identification of palindromic regions1. The symbols have been designed to underscore key biochemical properties of the nucleotides, trivialize the derivation and complementation of ambiguity characters, and support the visualization of sequence polymorphisms. The notation has five core components.
- Mnemonics. Ambiscript symbols use prominent typographic features to highlight key biochemical aspects of the nucleic acids they represent.
- Complementation. Complementary nucleotides are encoded by the rotationally related ambigraphic character, making it is possible to complement genetic sequences by simply rotating the text 180°.
- Palindromes. Character symmetries also make it easy to locate palindromic regions in Ambiscript encoded genetic sequences.
- Legibility. Distinctive ascenders and descenders facilitate the identification of nucleotide polymorphisms in multiple sequence alignments. An ultra-bold version of the Ambiscript notation further enhances sequence legibility.
- Ambiguity. Ambiguity characters are easily constructed and complemented by overlaying the symbols for the constituent bases.
We’ve designed a set of Truetype fonts to enable use of the notation in common word processor applications.
1 Rozak DA, Rozak AJ. 2008. Simplicity, function, and legibility in an enhanced ambirgaphic nucleic acid notation. BioTechniques 44(6):811-813. (PMID: 18476835)