Ambiscript uses prominent ascenders and descenders to enhance legibility
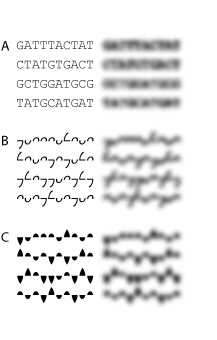
Since the early days of DNA analysis, researchers have pursued graphic solutions to support the visual analysis of genetic data1-4. In many cases, these techniques used size, color, shape, and vertical offsets of nucleic acid notations to highlight polymorphisms in multiple sequence alignments.
Ambiscript similarly exploits distinctive shapes offsets to facilitate visual review of nucleic acid sequences. Influenced chiefly by Tinker’s study of the Roman alphabet5, Ambiscript uses contrasting ascenders and descenders to support visual differentiation of the genetic symbols. The blurred text in the examples on the right illustrates the value of these external projections in discerning textually elements.
The figure on the right show-cases two versions of the Ambiscript font. The first font (Example B), was designed to accommodate those situations in which hand written sequences are called for. Indeed, this standard version of the Ambiscript notation boasts an average stroke per character (ASPC) of 2.50 compared to the 3.75 ASPC of the IUPAC notation.
Example C illustrates an ultra-bold Ambiscript font, whose filled-in contours enhance sequence legibility at the expense of writability. Both the normal and ultra-bold Ambiscript are available for free as downloadable Truetype fonts.
1 Cowin JE, Jellis CH, Rickwood D. 1986. A new method of representing DNA sequences which combines ease of visual analysis with machine readability. Nucleic Acids Res. 14:509-515. (PMID: 3003680)
2 Jarvius J, Landegren U. 2006. DNA Skyline: fonts to facilitate visual inspection of nucleic acid sequences. BioTechniques 40:740. (PMID: 16774117)
3 Schneider TD, Stephens RM. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097-6100. (PMID: 2172928)
4 Zimmerman PA, Spell ML, Rawls J, Unnasch TR. 1991. Transformation of DNA sequence data into geometric symbols. BioTechniques 11:50-52. (PMID: 1954017)
5 Tinker MA. 1963. Legibility of Print. Iowa State University Press, Ames, IA.